Enhanced False Discovery Rate (EFDR) tutorial
Andrew Zammit Mangion
Introduction
Enhanced False Discovery Rate (EFDR) is a nonparameteric hypothesis testing procedure used for anomaly detection in noisy spatial signals. The method, published by Shen et al. (2002), extends standard multiple hypothesis testing approaches (for example those employing the Bonferroni correction, or the standard false discovery rate (FDR) approach) by reducing the number of tests carried out. A reduced number of tests results in an approach which has more power (i.e. a lower Type II error), and hence an increased ability to detect anomalies. EFDR has proven to be vastly superior to the Bonferroni correction and superior to the standard FDR: the interested reader is referred to Shen et al. (2002) for a detailed study.
The purpose of this tutorial is to outline the basics of EFDR and demonstrate the use of the R package, EFDR, which can implement EFDR together with other basic hypothesis testing methods commonly used with spatial signals.
Underlying Theory
The ‘enhanced’ in EFDR is used to emphasise the increased power of the method over other conventional methods, brought about by reducing the number of hypothesis tests carried out when analysing a spatial signal. The number of tests carried out is reduced in two ways. First, the image is converted into the wavelet domain through a discrete wavelet transform (DWT); since the number of coefficients in wavelet space is generally chosen to be less than the number of pixels in the image, less tests are carried out. Second, tests are only carried out on some wavelet coefficients whose neighbours have relatively large wavelet coefficients.
The size of the wavelet neighbourhood  is determined by the user, while the definition of what constitutes a neighbour is determined by a metric which is fixed. This metric takes into consideration the (i) resolution, (ii) orientation, and (iii) spatial position of the wavelet. Wavelets at the same resolution are considered closer to each other than wavelets across resolutions. The same holds for orientation and spatial position. For more details on the distance metric employed, refer to Shen et al. (2002). In practice, the neighbours of wavelet
is determined by the user, while the definition of what constitutes a neighbour is determined by a metric which is fixed. This metric takes into consideration the (i) resolution, (ii) orientation, and (iii) spatial position of the wavelet. Wavelets at the same resolution are considered closer to each other than wavelets across resolutions. The same holds for orientation and spatial position. For more details on the distance metric employed, refer to Shen et al. (2002). In practice, the neighbours of wavelet 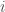 are found by calculating the distance to every other wavelet; the
are found by calculating the distance to every other wavelet; the  closest wavelets are then assigned as neighbours of the
closest wavelets are then assigned as neighbours of the 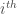 wavelet.
wavelet.
If there are 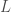 wavelets, then only
wavelets, then only  wavelets are tested, and usually
wavelets are tested, and usually 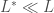 . A key difficulty is finding a suitable
. A key difficulty is finding a suitable  such that the Type I error rate is controlled at some pre-specified level
such that the Type I error rate is controlled at some pre-specified level 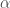 . One such method, which is proved to control the Type I error in the Appendix of Shen et al. (2002), is found by minimising a cost function which penalises for a large
. One such method, which is proved to control the Type I error in the Appendix of Shen et al. (2002), is found by minimising a cost function which penalises for a large  . The penalisation term requires the evaluation of an integral using Monte Carlo methods.
. The penalisation term requires the evaluation of an integral using Monte Carlo methods.
The EFDR R package: A toy study
As apparent from the previous section, EFDR is a multi-faceted appoach requiring wavelet transforms, neighbourhood selection in a wavelet domain and the evaluation of an optimal  . The package
. The package EFDR is designed to facilitate the implementation of EFDR, and uses computationally-efficient parallel methods to render manageable what is naively a highly computationally-intensive approach to false-discovery-rate detection. The package is loaded in R as follows. We will also load the package fields for plotting purposes:
library(EFDR)
library(fields)
Next, the user has to specify an image on which to carry out the analysis. The image needs to be an 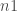 by
by  matrix where both
matrix where both 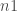 and $latex n2$ are integer powers of 2 (we also provide functions to deal with data which do not lie on a regular grid through the function
and $latex n2$ are integer powers of 2 (we also provide functions to deal with data which do not lie on a regular grid through the function regrid. This will be discussed later on in the tutorial). To get up and running, we have provided a function test_image which generates images similar to those analysed in Shen et al. (2002) with the package. In the following example we generate a 64 by 64 image and add some noise to it before plotting it (a filled circle embedded in Gaussian noise).
n = 64
Z <- test_image(n1=n)$z
Z_noisy <- Z + 0.2*rnorm(n^2)
fields::image.plot(Z,zlim=c(-0.5,1.5),asp=1)

fields::image.plot(Z_noisy,zlim=c(-0.5,1.5),asp=1)

Next, the user needs to provide some essential parameters. These are
alpha: significance levelwf: wavelet name (typically “la8”)J: number of multi-level resolutions (for a 64 x 64, 3 is sufficient)b: number of neighbours to consider with EFDRn.hyp: numeric vector of test counts, from which the optimal number of tests,  , will be found. Setting this to a scalar fixes the number of tests a priori
, will be found. Setting this to a scalar fixes the number of tests a prioriiteration: number of iterations in the Monte Carlo integration described earlierparallel: number of cores to use
alpha = 0.05 # 5% significant level
wf = "la8" # wavelet name
J = 3 # 3 resolutions
b = 11 # 11 neighbours for EFDR tests
n.hyp = c(5,10,20,30,40,60,80,100,400,800,1200,2400,4096) # find optimal number of tests in
# EFDR from this list
iteration = 100 # number of iterations in MC run when finding optimal n in n.hyp
idp = 0.5 # inverse distance weighting power
nmax = 15 # maximum number of points to consider when carrying out inverse
# distance interpolation
parallel = parallel::detectCores()/2 # use half the number of available cores
set.seed(20) # same random seed for reproducibility
That’s it! Now we can run EFDR. For comparison purposes, the package also provides functions for implementing the Bonferroni, the standard FDR and the Largest Order Statistic method (which tests the coefficient associated with the most extreme p-value, and deems it as significant if it less than  in wavelet space. The interface to each of these methods is similar.
in wavelet space. The interface to each of these methods is similar.
m1 <- test.bonferroni(Z_noisy, wf=wf,J=J, alpha = alpha)
m2 <- test.fdr(Z_noisy, wf=wf,J=J, alpha = alpha)
m3 <- test.los(Z_noisy, wf=wf,J=J, alpha = alpha)
m4 <- test.efdr(Z_noisy, wf="la8",J=J, alpha = alpha, n.hyp = n.hyp,
b=b,iteration=iteration,parallel = parallel)
## Starting EFDR ...
## ... Finding neighbours ...
## ... Estimating the EFDR optimal number of tests ...
The functions provide several results, mostly for diagnostic purposes, in a list (see documentation for details). The result of most interest is the image which contains only the ‘extreme’ wavelet coefficients, stored in the field Z. These can be plotted using standard plotting functions.
par(mfrow=c(2,2))
fields::image.plot(m1$Z,main = "Bonferroni",zlim=c(-1.5,2),asp=1)
fields::image.plot(m2$Z,main = "FDR",zlim=c(-1.5,2),asp=1)
fields::image.plot(m3$Z,main = "LOS",zlim=c(-1.5,2),asp=1)
fields::image.plot(m4$Z,main = "EFDR",zlim=c(-1.5,2),asp=1)

To compute the power of the test (which we can, since we know the truth), we can use the functions wav_th and fdrpower. The first determines which wavelength coefficients are ‘significant’ by seeing which, in the raw signal, exceed a given threshold. The second function determines which of these wavelet coefficients were correctly found to be rejected under the null hypothesis that the coefficient is zero
sig <- wav_th(Z,wf=wf,J=J,th=1)
cat(paste0("Power of Bonferroni test: ",fdrpower(sig,m1$reject_coeff)),
paste0("Power of FDR test: ",fdrpower(sig,m2$reject_coeff)),
paste0("Power of LOS test: ",fdrpower(sig,m3$reject_coeff)),
paste0("Power of EFDR test: ",fdrpower(sig,m4$reject_coeff)),sep="\n")
## Power of Bonferroni test: 0.85
## Power of FDR test: 0.95
## Power of LOS test: 0.05
## Power of EFDR test: 0.95
Another tool delivered with the package is a diagnostic table containing the number of false positives, false negatives, true negatives and true positives.
### Bonferroni diagnostic table:
diagnostic.table(sig,m1$reject_coeff,n = n^2)
## Real Negative Real Positive
## Diagnostic Negative 4073 3
## Diagnostic Positive 3 17
### FDR diagnostic table:
diagnostic.table(sig,m2$reject_coeff,n = n^2)
## Real Negative Real Positive
## Diagnostic Negative 4064 12
## Diagnostic Positive 1 19
### LOS diagnostic table:
diagnostic.table(sig,m3$reject_coeff,n = n^2)
## Real Negative Real Positive
## Diagnostic Negative 4076 0
## Diagnostic Positive 19 1
### EFDR diagnostic table:
diagnostic.table(sig,m4$reject_coeff,n = n^2)
## Real Negative Real Positive
## Diagnostic Negative 4062 14
## Diagnostic Positive 1 19
Several useful diagnostic measures can be computed from this table. In particular, define
 : Number of true negatives
: Number of true negatives : Number of false positives
: Number of false positives : Number of false negatives
: Number of false negatives : Number of true positives
: Number of true positives
Then, the false discovery rate (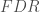 ), false nondiscovery rate (
), false nondiscovery rate ( ), specificity (
), specificity ( ) and sensitivity (
) and sensitivity ( ) can be computed as
) can be computed as




This example considered a relatively ‘clean’ image. The real power of EFDR becomes apparent when analysing very noisy images, where the signal is not even visible from a casual inspection and hence having a powerful testing procedure becomes important. In the following we take the same signal, but this time add on a considerable amount of noise before running the aforementioned tests.
set.seed(1) # for reproducibility
Z <- test_image(n1=n)$z
Z_noisy <- Z + 2*rnorm(n^2)
fields::image.plot(Z_noisy)

m1 <- test.bonferroni(Z_noisy, wf=wf,J=J, alpha = alpha)
m2 <- test.fdr(Z_noisy, wf=wf,J=J, alpha = alpha)
m3 <- test.los(Z_noisy, wf=wf,J=J, alpha = alpha)
m4 <- test.efdr(Z_noisy, wf="la8",J=J, alpha = alpha, n.hyp = n.hyp,
b=b,iteration=iteration,parallel = parallel)
## Starting EFDR ...
## ... Finding neighbours ...
## ... Estimating the EFDR optimal number of tests ...
par(mfrow=c(2,2))
fields::image.plot(m1$Z,main = "Bonferroni", zlim=c(-1.5,2),asp=1)
fields::image.plot(m2$Z,main = "FDR", zlim=c(-1.5,2),asp=1)
fields::image.plot(m3$Z,main = "LOS", zlim=c(-1.5,2),asp=1)
fields::image.plot(m4$Z,main = "EFDR", zlim=c(-1.5,2))

sig <- wav_th(Z,wf=wf,J=J,th=1)
cat(paste0("Power of Bonferroni test: ",fdrpower(sig,m1$reject_coeff)),
paste0("Power of FDR test: ",fdrpower(sig,m2$reject_coeff)),
paste0("Power of LOS test: ",fdrpower(sig,m3$reject_coeff)),
paste0("Power of EFDR test: ",fdrpower(sig,m4$reject_coeff)),sep="\n")
## Power of Bonferroni test: 0
## Power of FDR test: 0
## Power of LOS test: 0
## Power of EFDR test: 0.3
In fact, in this case no method detects an anomaly except for EFDR. Note that some spurious signals are detected, this is a result of an larger Type I error than the other methods, but recall that EFDR’s Type I error rate is still constrained to be less than 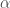 .
.
A real example
In this example we implement all the above tests on data from the Atmospheric Infrared Sounder (AIRS). Data are available from the NIASRA data portal.
As a simple case study, we take CO2 data from the first three days in these data (01 – 03 May 2003) and compare these to data in the second three days (4 – 6 May 2003) in the conterminus United States and Canada. The change in CO2 in these periods is an indication of regional fluxes and we are interested in where the flux is significantly different from zero. This satellite data is irregular in space; it therefore needs to be first regridded and interpolated onto an image of appropriate size.
First, load the dataset using RCurl:
library(RCurl)
airs.raw <- read.table(textConnection(getURL(
"http://hpc.niasra.uow.edu.au/ckan/dataset/6ddf61bb-2f9a-424a-8775-e23ebaa55afb/resource/e1206e1c-9dca-42b6-8455-3cdd98c6a943/download/airs2003may116.csv"
)),header=T,sep=",")
Now we will concentrate on North America, and change the variable names into the appropriate x-y-z notation required to regrid the data. For this we use some handy functions from the dplyr package.
library(dplyr)
airs.raw <- airs.raw %>%
subset(lat > 14 & lat < 66 & lon > -145 & lon < -52) %>%
mutate(x=lon,y=lat,z=co2avgret) %>%
select(day,x,y,z)
For plotting we use the ggplot2 package which includes the world coastline. Below we plot all the data in the first three days of the dataset over North America.
library(ggplot2)
NorthAmerica <- map_data("world") %>%
subset(region %in% c("USA","Canada","Mexico"))
par(pin = c(5,1))
ggplot() +
geom_point(data=subset(airs.raw,day%in% 1:3),aes(x=x,y=y,colour=z)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_colour_gradient(low="yellow",high="blue") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66))

In order to put this data frame into an image or matrix format, we use EFDR::regrid. Regrid requires three parameters:
n1: the length (in pixels) of the imagen2: (optional) the height (in pixels) of the imageidp: the inverse distance power used in the inverse distance weighting interpolation function used to fill in the gaps (e.g. idw = 2)nmax: the maximum number of neighbours to consider when carrying out the interpolation (for computational efficiency this should be low, around 8).
In the following example we use an inverse squared distance weighting and re-grid the AIRS CO2 data from 1–3 May 2003 into a 256 x 128 image.
idp = 0.5
n1 = 256
n2 = 128
airs.interp1 <- airs.raw %>%
subset(day %in% 1:3) %>%
select(-day) %>%
regrid(n1=n1,n2=n2,idp= idp,nmax = nmax)
## [inverse distance weighted interpolation]
# idp is the inverse distance power and
# nmax is the number of neighbours to consider
ggplot() +
geom_tile(data=airs.interp1,aes(x=x,y=y,fill=z)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient(low="yellow",high="blue") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66))

We are interested in changes in CO2 levels. Below we produce an image of CO2 fluxes by repeating the regridding on the AIRS CO2 data from 4–6 May 2003 and then finding the difference between the two images
airs.interp2 <- airs.raw %>%
subset(day %in% 4:6) %>%
select(-day) %>%
regrid(n1=n1,n2=n2,idp= idp,nmax = nmax)
## [inverse distance weighted interpolation]
airs.interp <- airs.interp2 %>%
mutate(z = airs.interp1$z - airs.interp2$z)
The differenced image looks as follows
ggplot() +
geom_tile(data=airs.interp,aes(x=x,y=y,fill=z)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient2(low="blue",high="red", mid="#FFFFCC") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66))

We now convert the data frame to an image matrix (using the package function df.to.mat) and then carry out the wavelet-domain tests, including EFDR.
Z <- df.to.mat(airs.interp)
m1 <- test.bonferroni(Z, wf=wf,J=J, alpha = alpha)
m2 <- test.fdr(Z, wf=wf,J=J, alpha = alpha)
m3 <- test.los(Z, wf=wf,J=J, alpha = alpha)
m4 <- test.efdr(Z, wf="la8",J=J, alpha = alpha, n.hyp = n.hyp,
b=b,iteration=iteration,parallel = parallel)
## Starting EFDR ...
## ... Finding neighbours ...
## ... Estimating the EFDR optimal number of tests ...
For plotting we re-arrange the data into data frames and then repeat the above. For gridding ggplot objects we will use the gridExtra package.
library(gridExtra)
airs.interp <- airs.interp %>%
mutate(m1 = c(m1$Z),
m2 = c(m2$Z),
m3 = c(m3$Z),
m4 = c(m4$Z))
g1 <- ggplot() +
geom_tile(data=airs.interp,aes(x=x,y=y,fill=m1)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient2(low="blue",high="red", mid="#FFFFCC") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66)) + ggtitle("Bonferroni")
g2 <- ggplot() +
geom_tile(data=airs.interp,aes(x=x,y=y,fill=m2)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient2(low="blue",high="red", mid="#FFFFCC") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66)) + ggtitle("FDR")
g3 <- ggplot() +
geom_tile(data=airs.interp,aes(x=x,y=y,fill=m3)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient2(low="blue",high="red", mid="#FFFFCC") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66)) + ggtitle("LOS")
g4 <- ggplot() +
geom_tile(data=airs.interp,aes(x=x,y=y,fill=m4)) +
geom_path(data=NorthAmerica,aes(x=long,y=lat,group=group)) +
scale_fill_gradient2(low="blue",high="red", mid="#FFFFCC") +
coord_fixed(xlim=c(-145,-52),ylim=c(14,66)) + ggtitle("EFDR")
gridExtra::grid.arrange(g1, g2, g3, g4, ncol=2)

Once again, EFDR is seen to detect more anomalies than all other methods. In this case both LOS and the standard Bonferroni correction do not detect anomalies due to a poor power. Another example demonstrating the increased power of EFDR using a temperature difference dataset can be found in Shen et al. (2002).
Reference
Shen, Xiaotong, Hsin-Cheng Huang, and Noel Cressie. “Nonparametric hypothesis testing for a spatial signal.” Journal of the American Statistical Association 97.460 (2002): 1122-1140.














